Example with IPD trial data
Valerie Aponte Ribero and Javier Sanchez Alvarez
December 15, 2025
Source:vignettes/articles/example_ipd.Rmd
example_ipd.RmdIntroduction
This document runs a discrete event simulation model using simulated individual patient data (IPD) to show how the functions can be used to generate a model when IPD from a trial is available.
Packages and main options
Model concept
All patients start in the progression-free state and may move to the progressed state. At any point in time they can die, depending on the risk of each disease stage. Patients may also experience a disease event which accelerates progression.
Generate dummy IPD trial data and fit survival models
The dummy IPD trial data was generated below from using the
sim_adtte() function from the flexsurvPlus
package. Parametric survival models are fit to the dummy OS and TTP IPD.
We are using the flexsurv package to fit the parametric survival
models.
#Generate dummy IPD
tte.df <- WARDEN::tte.df
#Change data frame to wide format
tte.df <- tte.df %>% select(-PARAM) %>% pivot_wider(names_from = PARAMCD, values_from = c(AVAL,CNSR))
#Derive Time to Progression Variable from OS and PFS
tte.df <- tte.df %>% mutate(
AVAL_TTP = AVAL_PFS,
CNSR_TTP = ifelse(AVAL_PFS == AVAL_OS & CNSR_PFS==0 & CNSR_OS==0,1,CNSR_PFS),
Event_OS = 1-CNSR_OS,
Event_PFS = 1-CNSR_PFS,
Event_TTP = 1-CNSR_TTP
)
#Add baseline characteristics (sex and age) to time to event data
IPD <- tte.df %>% mutate(
SEX = rbinom(500,1,0.5),
AGE = rnorm(500,60,8)
)
#Plot simulated OS and TTP curves
#Overall survival
km.est.OS <- survfit(Surv(AVAL_OS/365.25, Event_OS) ~ ARMCD, data = IPD) #KM curve
OS.fit <- flexsurvreg(formula = Surv(AVAL_OS/365.25, Event_OS) ~ ARMCD, data = IPD, dist = "Weibull") #Fit Weibull model to the OS data
OS.fit
#> Call:
#> flexsurvreg(formula = Surv(AVAL_OS/365.25, Event_OS) ~ ARMCD,
#> data = IPD, dist = "Weibull")
#>
#> Estimates:
#> data mean est L95% U95% se exp(est) L95% U95%
#> shape NA 1.1346 0.9764 1.3185 0.0870 NA NA NA
#> scale NA 3.1523 2.5170 3.9479 0.3620 NA NA NA
#> ARMCDB 0.5000 0.3066 0.0183 0.5949 0.1471 1.3588 1.0185 1.8129
#>
#> N = 500, Events: 150, Censored: 350
#> Total time at risk: 623.0253
#> Log-likelihood = -360.0964, df = 3
#> AIC = 726.1929
ggsurvplot(OS.fit, title="Overall survival",
legend.labs = c("Reference","Intervention"),
risk.table = TRUE)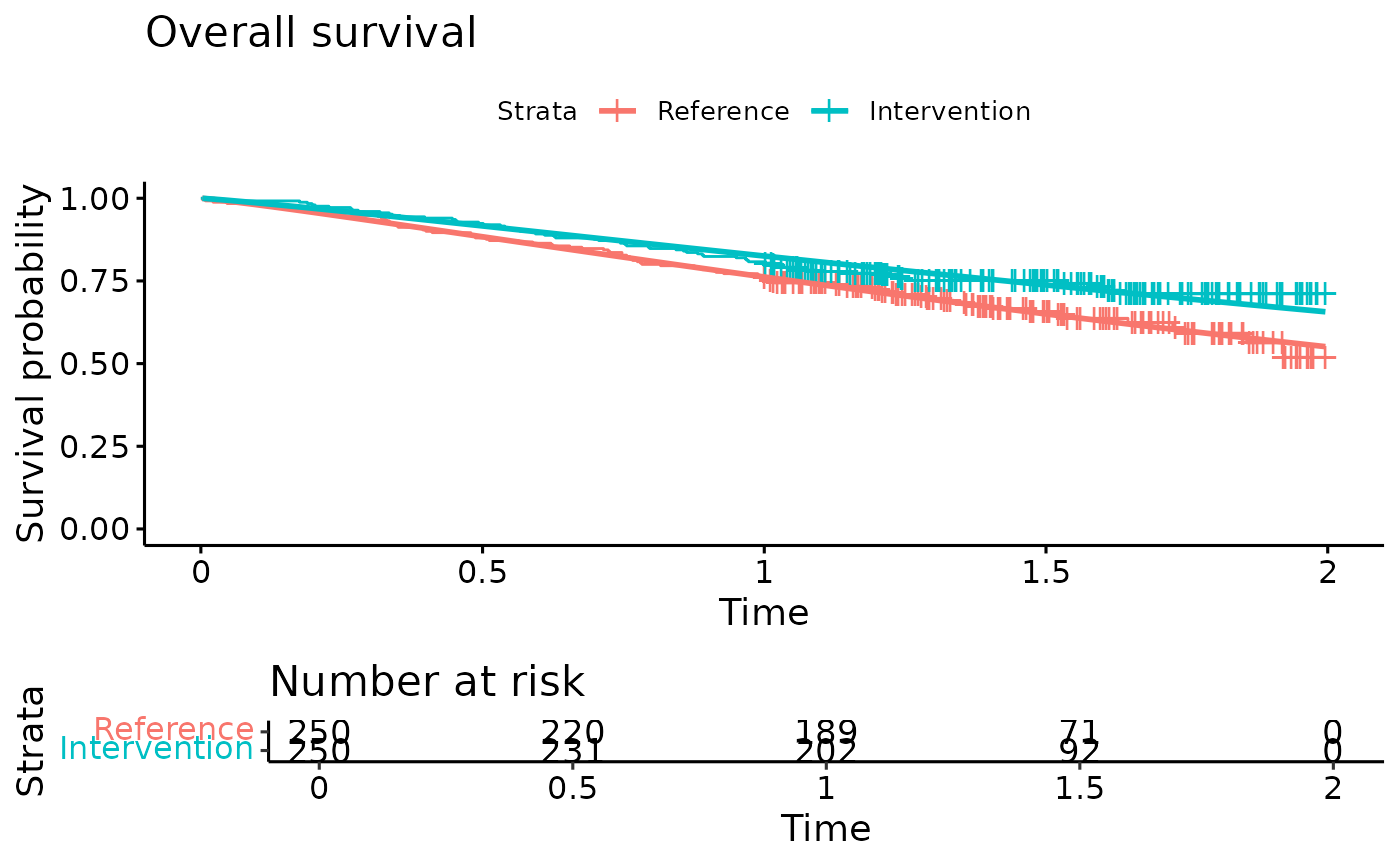
#Time to progression
km.est.TTP <- survfit(Surv(AVAL_TTP/365.25, Event_TTP) ~ ARMCD, data = IPD) #KM curve
TTP.fit <- flexsurvreg(formula = Surv(AVAL_TTP/365.25, Event_TTP) ~ ARMCD, data = IPD, dist = "Weibull") #Fit Weibull model to the TTP data
TTP.fit
#> Call:
#> flexsurvreg(formula = Surv(AVAL_TTP/365.25, Event_TTP) ~ ARMCD,
#> data = IPD, dist = "Weibull")
#>
#> Estimates:
#> data mean est L95% U95% se exp(est) L95% U95%
#> shape NA 1.3757 1.2590 1.5033 0.0622 NA NA NA
#> scale NA 0.5865 0.5310 0.6478 0.0297 NA NA NA
#> ARMCDB 0.5000 1.0992 0.9277 1.2707 0.0875 3.0016 2.5286 3.5632
#>
#> N = 500, Events: 328, Censored: 172
#> Total time at risk: 358.9897
#> Log-likelihood = -269.0387, df = 3
#> AIC = 544.0774
ggsurvplot(TTP.fit, title="Time to progression",
legend.labs = c("Reference","Intervention"),
risk.table = TRUE)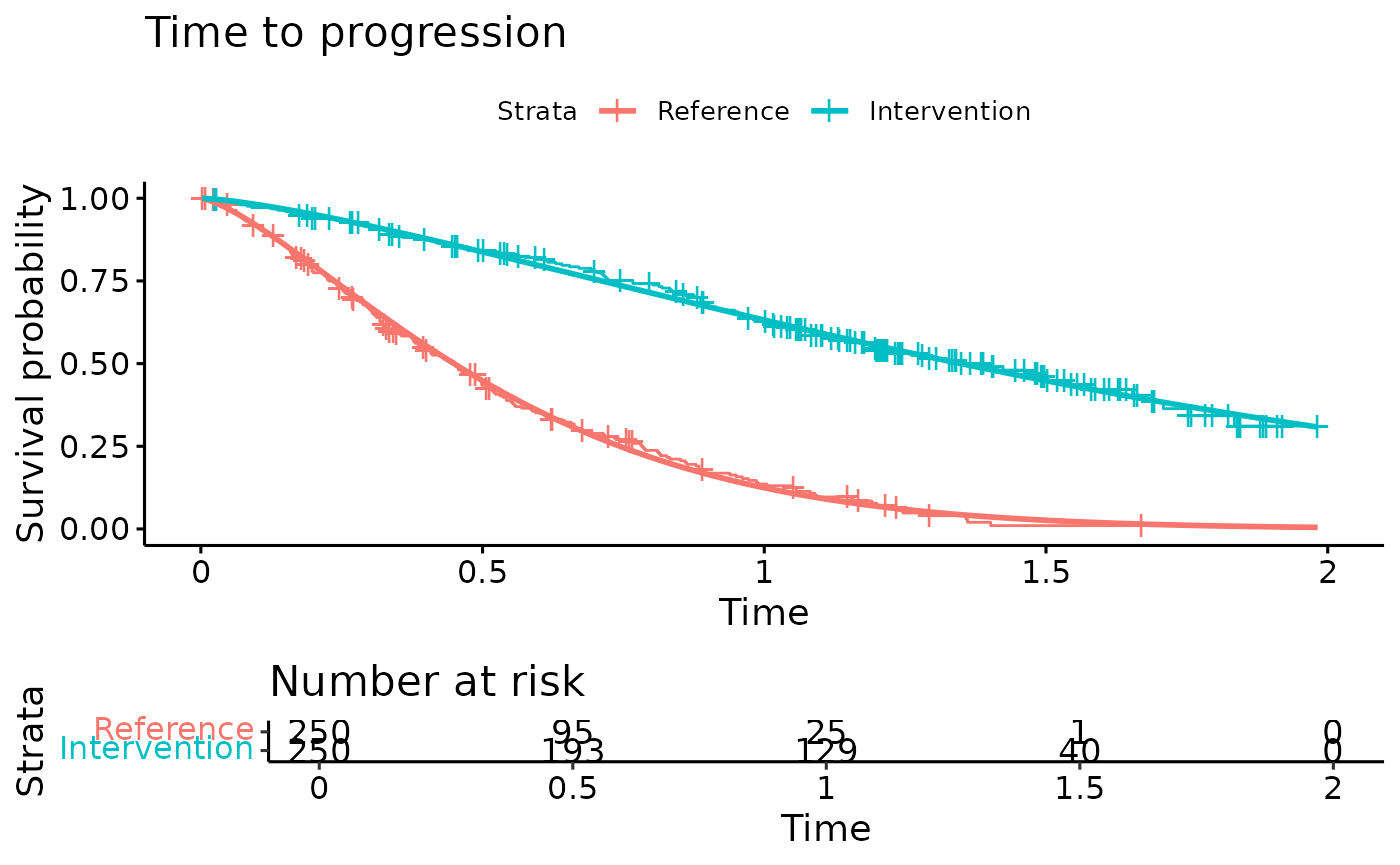
Define DES model inputs
Inputs and variables that will be used in the model are defined
below. We can define inputs that are common to all patients
(common_all_inputs) within a simulation, inputs that are
unique to a patient independently of the treatment (e.g. natural death,
defined in common_pt_inputs), and inputs that are unique to
that patient and that treatment (unique_pt_inputs). Items
can be included through the add_item() function, and can be
used in subsequent items. All these inputs are generated before the
events and the reaction to events are executed. Furthermore, the program
first executes common_all_inputs, then
common_pt_inputs and then unique_pt_inputs. So
one could use the items generated in common_all_inputs in
unique_pt_inputs.
#Define variables that do not change on any patient or intervention loop
common_all_inputs <- add_item(
#Parameters from the survival models
OS.scale = as.numeric(OS.fit$coef[2]),
OS.shape = as.numeric(OS.fit$coef[1]),
OS.coef.int = as.numeric(OS.fit$coef[3]), #Intervention effect
TTP.scale = as.numeric(TTP.fit$coef[2]),
TTP.shape = as.numeric(TTP.fit$coef[1]),
TTP.coef.int = as.numeric(TTP.fit$coef[3]), #Intervention effect
#Utilities
util.PFS = 0.6, #Utility while in progression-free state
util.PPS = 0.4, #Utility while in progressed state
disutil.PAE = -0.02, #One-off disutility of progression-accelerating event
#Costs
cost.drug.int = 85000, #Annual intervention cost
cost.drug.ref = 29000, #Annual cost of reference treatment
cost.admin.SC = 150, #Unit cost for each SC administration
cost.admin.oral = 300, #One-off cost for oral administration
cost.dm.PFS = 3000, #Annual disease-management cost in progression-free state
cost.dm.PPS = 5000, #Annual disease-management cost in progressed state
cost.ae.int = 2200, #Annual adverse event costs for intervention
cost.ae.ref = 1400 #Annual adverse event costs for reference treatment
)
#Define variables that do not change as we loop through interventions for a patient
common_pt_inputs <- add_item(
#Patient baseline characteristics
Sex = as.numeric(IPD[i,"SEX"]), #Record sex of individual patient. 0 = Female; 1 =Male
BLAge = as.numeric(IPD[i,"AGE"]), #Record patient age at baseline
#Draw time to non-disease related death from a conditional Gompertz distribution
nat.death = rcond_gompertz(1,shape=if(Sex==1){0.102}else{0.115},
rate=if(Sex==1){0.000016}else{0.0000041},
lower_bound = BLAge) # Baseline Age in years
)
#Define variables that change as we loop through treatments for each patient.
unique_pt_inputs <- add_item(
fl.int = 0, #Flag to determine if patient is on intervention. Initialized as 0, but will be changed to current arm in the Start event.
fl.prog = 0, #Flag to determine if patient has progressed. All patients start progression-free
fl.ontx = 1, #Flag to determine if patient is on treatment. All patients start on treatment
fl.PAE = 0, #Flag to determine if progression-accelerating event occurred
pfs.time = NA, #Recording of time at progression
q_default = ifelse(fl.prog == 0, util.PFS, util.PPS),
q_default_inst = 0,
c_default = ifelse(fl.prog == 0,cost.dm.PFS,cost.dm.PPS) + if(arm=="int"){(cost.drug.int + cost.admin.SC * 12 + cost.ae.int) * fl.ontx}else{cost.drug.ref + cost.ae.ref},
c_default_inst = 0
)Events
Add Initial Events
We define now the possible events that can occur for the intervention
and reference arm respectively using the add_tte()
function. Only patients in the intervention arm can have a treatment
discontinuation, while patients in both arms can have a progression,
progression-accelerating and death event. The seed argument is being
used in the draw_tte() function which uses the
i item to ensure that event times specific to each patient
can be replicated and updated at later time points.
init_event_list <-
#Events applicable to intervention
add_tte(arm=c("int","ref"),
evts = c("Start",
"TxDisc",
"Progression",
"PAE",
"Death"),
input={
Start <- 0
Progression <- draw_tte(1,'weibull',coef1=TTP.shape, coef2= TTP.scale + ifelse(arm=="int",TTP.coef.int,0),seed = as.numeric(paste0(1,i,simulation)))
TxDisc <- Inf #Treatment discontinuation will occur at progression
Death <- min(draw_tte(1,'weibull',coef1=OS.shape, coef2= OS.scale + ifelse(arm=="int",OS.coef.int,0), seed = as.numeric(paste0(42,i,simulation))), nat.death) #Death occurs at earliest of disease-related death or non-disease-related death
PAE <- draw_tte(1,'exp',coef1=-log(1-ifelse(arm=="int",0.05,0.15))) #Occurrence of the progression-accelerating event has a 5% or 15% probability for the intervention arm
})Add Event Reactions
Reactions for each individual event are defined in the following
using the add_reactevt() function. Patients in the
intervention arm discontinue treatment at progression. Occurrence of the
progression-accelerated event results in an earlier progression (if it
has not occurred yet). Note the use of the seed argument in the
draw_tte() function which ensures that the same seed is
being used in the original and the updated draw of the time to
progression.
evt_react_list <-
add_reactevt(name_evt = "Start",
input = {
fl.int = ifelse(arm=="int",1,0)
q_default = ifelse(fl.prog == 0, util.PFS, util.PPS)
c_default = ifelse(fl.prog == 0,cost.dm.PFS,cost.dm.PPS) + if(arm=="int"){(cost.drug.int + cost.admin.SC * 12 + cost.ae.int) * fl.ontx}else{cost.drug.ref + cost.ae.ref}
c_default_inst = cost.admin.oral
}) %>%
add_reactevt(name_evt = "TxDisc",
input = {
q_default = ifelse(fl.prog == 0, util.PFS, util.PPS)
c_default = ifelse(fl.prog == 0,cost.dm.PFS,cost.dm.PPS) + if(arm=="int"){(cost.drug.int + cost.admin.SC * 12 + cost.ae.int) * fl.ontx}else{cost.drug.ref + cost.ae.ref}
fl.ontx = 0
}) %>%
add_reactevt(name_evt = "Progression",
input = {
q_default = ifelse(fl.prog == 0, util.PFS, util.PPS)
c_default = ifelse(fl.prog == 0,cost.dm.PFS,cost.dm.PPS) + if(arm=="int"){(cost.drug.int + cost.admin.SC * 12 + cost.ae.int) * fl.ontx}else{cost.drug.ref + cost.ae.ref}
pfs.time=curtime
fl.prog= 1
if(arm=="int"){modify_event(c("TxDisc" = curtime))} #Trigger treatment discontinuation at progression
}) %>%
add_reactevt(name_evt = "Death",
input = {
q_default = ifelse(fl.prog == 0, util.PFS, util.PPS)
c_default = ifelse(fl.prog == 0,cost.dm.PFS,cost.dm.PPS) + if(arm=="int"){(cost.drug.int + cost.admin.SC * 12 + cost.ae.int) * fl.ontx}else{cost.drug.ref + cost.ae.ref}
curtime = Inf
}) %>%
add_reactevt(name_evt = "PAE",
input = {
fl.PAE = 1
q_default = ifelse(fl.prog == 0, util.PFS, util.PPS)
q_default_inst = disutil.PAE
c_default = ifelse(fl.prog == 0,cost.dm.PFS,cost.dm.PPS) + if(arm=="int"){(cost.drug.int + cost.admin.SC * 12 + cost.ae.int) * fl.ontx}else{cost.drug.ref + cost.ae.ref}
if(fl.prog == 0){ #Event only accelerates progression if progression has not occurred yet
modify_event(c(
"Progression"=max(draw_tte(1,'weibull',coef1=TTP.shape, coef2= TTP.scale + TTP.coef.int*fl.int, beta_tx = 1.2, seed = as.numeric(paste0(1,i,simulation))),curtime))) #Occurrence of event accelerates progression by a factor of 1.2
}
})Model
Model Execution
The model is executed with the event reactions and inputs previously defined for each patient in the simulated data set.
#Logic is: per patient, per intervention, per event, react to that event.
results <- run_sim(
npats=as.numeric(nrow(IPD)), # Simulating the number of patients for which we have IPD
n_sim=1, # We run all patients once (per treatment)
psa_bool = FALSE, # No PSA for this example
arm_list = c("int", "ref"),
common_all_inputs = common_all_inputs,
common_pt_inputs = common_pt_inputs,
unique_pt_inputs = unique_pt_inputs,
init_event_list = init_event_list,
evt_react_list = evt_react_list,
util_ongoing_list = util_ongoing,
util_instant_list = util_instant,
cost_ongoing_list = cost_ongoing,
cost_instant_list = cost_instant,
input_out = c("BLAge","Sex","nat.death","pfs.time")
)
#> Analysis number: 1
#> Simulation number: 1
#> Time to run simulation 1: 0.67s
#> Time to run analysis 1: 0.67s
#> Total time to run: 0.68s
#> Simulation finalized;Post-processing of Model Outputs
Summary of Results
Once the model has been run, we can use the results and summarize
them using the summary_results_det to print the results of
the deterministic case. The individual patient data generated by the
simulation is recorded in the psa_ipd object.
summary_results_det(results[[1]][[1]]) #will print the last simulation!
#> int ref
#> costs 261251.76 94361.19
#> dcosts 0.00 166890.57
#> lys 3.60 2.75
#> dlys 0.00 0.85
#> qalys 1.71 1.41
#> dqalys 0.00 0.30
#> ICER NA 195784.55
#> ICUR NA 563768.08
#> INMB NA -152089.22
#> costs_undisc 286099.08 101986.36
#> dcosts_undisc 0.00 184112.72
#> lys_undisc 3.99 2.97
#> dlys_undisc 0.00 1.02
#> qalys_undisc 1.88 1.52
#> dqalys_undisc 0.00 0.35
#> ICER_undisc NA 180232.81
#> ICUR_undisc NA 518666.81
#> INMB_undisc NA -166364.07
#> BLAge 59.66 59.66
#> dBLAge 0.00 0.00
#> c_default 260951.76 94061.19
#> dc_default 0.00 166890.57
#> c_default_inst 300.00 300.00
#> dc_default_inst 0.00 0.00
#> c_default_inst_undisc 300.00 300.00
#> dc_default_inst_undisc 0.00 0.00
#> c_default_undisc 285799.08 101686.36
#> dc_default_undisc 0.00 184112.72
#> nat.death 24.33 24.33
#> dnat.death 0.00 0.00
#> pfs.time 1.61 0.58
#> dpfs.time 0.00 1.03
#> q_default 1.73 1.43
#> dq_default 0.00 0.30
#> q_default_inst -0.02 -0.02
#> dq_default_inst 0.00 0.00
#> q_default_inst_undisc -0.02 -0.02
#> dq_default_inst_undisc 0.00 0.00
#> q_default_undisc 1.89 1.54
#> dq_default_undisc 0.00 0.36
#> Sex 0.53 0.53
#> dSex 0.00 0.00
psa_ipd <- bind_rows(map(results[[1]], "merged_df"))
psa_ipd[1:10,] %>%
kable() %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))| evtname | evttime | prevtime | pat_id | arm | total_lys | total_qalys | total_costs | total_costs_undisc | total_qalys_undisc | total_lys_undisc | lys | qalys | costs | lys_undisc | qalys_undisc | costs_undisc | BLAge | Sex | nat.death | pfs.time | c_default | c_default_inst | q_default | q_default_inst | c_default_undisc | q_default_undisc | c_default_inst_undisc | q_default_inst_undisc | nexttime | simulation | sensitivity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Start | 0.0000000 | 0.000000 | 1 | int | 1.5055640 | 0.8841831 | 138811.88 | 141988.56 | 0.9040559 | 1.5400931 | 1.4288465 | 0.8573079 | 131753.8777 | 1.4598976 | 0.8759386 | 134610.5792 | 53.35829 | 0 | 30.07156 | NA | 131453.8777 | 300 | 0.8573079 | 0.0000000 | 134310.5792 | 0.8759386 | 300 | 0.00 | 1.4598976 | 1 | 1 |
| PAE | 1.4598976 | 0.000000 | 1 | int | 1.5055640 | 0.8841831 | 138811.88 | 141988.56 | 0.9040559 | 1.5400931 | 0.0767175 | 0.0268752 | 7058.0072 | 0.0801955 | 0.0281173 | 7377.9851 | 53.35829 | 0 | 30.07156 | NA | 7058.0072 | 0 | 0.0460305 | -0.0191553 | 7377.9851 | 0.0481173 | 0 | -0.02 | 1.5400931 | 1 | 1 |
| Death | 1.5400931 | 1.459898 | 1 | int | 1.5055640 | 0.8841831 | 138811.88 | 141988.56 | 0.9040559 | 1.5400931 | 0.0000000 | 0.0000000 | 0.0000 | 0.0000000 | 0.0000000 | 0.0000 | 53.35829 | 0 | 30.07156 | NA | 0.0000 | 0 | 0.0000000 | 0.0000000 | 0.0000 | 0.0000000 | 0 | 0.00 | 1.5400931 | 1 | 1 |
| Start | 0.0000000 | 0.000000 | 2 | int | 2.1750928 | 1.2858641 | 200408.54 | 207131.08 | 1.3288984 | 2.2481639 | 1.3675671 | 0.8205403 | 126116.1722 | 1.3959764 | 0.8375858 | 128729.8244 | 55.97126 | 1 | 36.09357 | NA | 125816.1722 | 300 | 0.8205403 | 0.0000000 | 128429.8244 | 0.8375858 | 300 | 0.00 | 1.3959764 | 1 | 1 |
| PAE | 1.3959764 | 0.000000 | 2 | int | 2.1750928 | 1.2858641 | 200408.54 | 207131.08 | 1.3288984 | 2.2481639 | 0.8075257 | 0.4653239 | 74292.3646 | 0.8521876 | 0.4913125 | 78401.2566 | 55.97126 | 1 | 36.09357 | NA | 74292.3646 | 0 | 0.4845154 | -0.0191915 | 78401.2566 | 0.5113125 | 0 | -0.02 | 2.2481639 | 1 | 1 |
| Death | 2.2481639 | 1.395976 | 2 | int | 2.1750928 | 1.2858641 | 200408.54 | 207131.08 | 1.3288984 | 2.2481639 | 0.0000000 | 0.0000000 | 0.0000 | 0.0000000 | 0.0000000 | 0.0000 | 55.97126 | 1 | 36.09357 | NA | 0.0000 | 0 | 0.0000000 | 0.0000000 | 0.0000 | 0.0000000 | 0 | 0.00 | 2.2481639 | 1 | 1 |
| Start | 0.0000000 | 0.000000 | 3 | int | 0.5544693 | 0.3326816 | 51311.17 | 51733.82 | 0.3354379 | 0.5590632 | 0.5544693 | 0.3326816 | 51311.1718 | 0.5590632 | 0.3354379 | 51733.8186 | 50.45087 | 1 | 25.36304 | NA | 51011.1718 | 300 | 0.3326816 | 0.0000000 | 51433.8186 | 0.3354379 | 300 | 0.00 | 0.5590632 | 1 | 1 |
| Death | 0.5590632 | 0.000000 | 3 | int | 0.5544693 | 0.3326816 | 51311.17 | 51733.82 | 0.3354379 | 0.5590632 | 0.0000000 | 0.0000000 | 0.0000 | 0.0000000 | 0.0000000 | 0.0000 | 50.45087 | 1 | 25.36304 | NA | 0.0000 | 0 | 0.0000000 | 0.0000000 | 0.0000 | 0.0000000 | 0 | 0.00 | 0.5590632 | 1 | 1 |
| Start | 0.0000000 | 0.000000 | 4 | int | 0.9496596 | 0.5503533 | 87668.68 | 88918.38 | 0.5579459 | 0.9632432 | 0.9431472 | 0.5658883 | 87069.5412 | 0.9565434 | 0.5739261 | 88301.9947 | 53.98621 | 0 | 37.38616 | NA | 86769.5412 | 300 | 0.5658883 | 0.0000000 | 88001.9947 | 0.5739261 | 300 | 0.00 | 0.9565434 | 1 | 1 |
| PAE | 0.9565434 | 0.000000 | 4 | int | 0.9496596 | 0.5503533 | 87668.68 | 88918.38 | 0.5579459 | 0.9632432 | 0.0065124 | -0.0155350 | 599.1403 | 0.0066998 | -0.0159801 | 616.3833 | 53.98621 | 0 | 37.38616 | NA | 599.1403 | 0 | 0.0039074 | -0.0194424 | 616.3833 | 0.0040199 | 0 | -0.02 | 0.9632432 | 1 | 1 |
We can also check what has been the absolute number of events per strategy.
| arm | evtname | n |
|---|---|---|
| int | Death | 500 |
| int | Start | 500 |
| int | PAE | 411 |
| int | Progression | 320 |
| int | TxDisc | 320 |
| ref | Death | 500 |
| ref | Start | 500 |
| ref | Progression | 441 |
| ref | PAE | 395 |
Plots
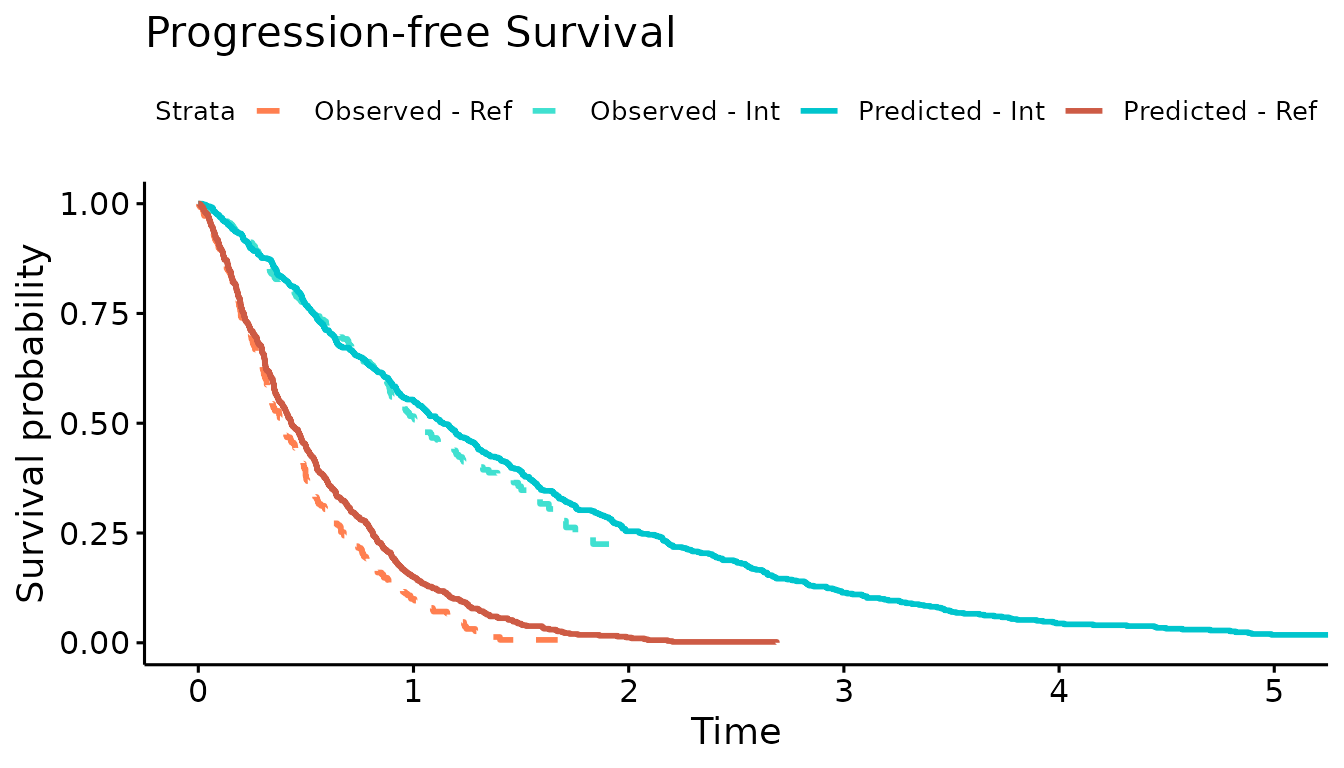
We now use the simulation output to plot the Kaplan-Meier curves of the simulated OS and PFS against the observed Kaplan-Meier curves. The simulated progression-free survival curve is lower than the observed due to the addition of the progression-accelerating event into the model.
#Overall survival
KM.death <- psa_ipd %>% filter(evtname=="Death") %>% mutate(Event = 1)
sim.km.OS <- survfit(Surv(evttime, Event) ~ arm, data = KM.death)
km.comb <- list(
Observed = km.est.OS,
Predicted = sim.km.OS
)
ggsurvplot(km.comb, combine = TRUE,
title="Overall Survival",
palette=c("coral","turquoise","turquoise3","coral3"),
legend.labs=c("Observed - Ref","Observed - Int","Predicted - Int","Predicted - Ref"),
linetype = c(2,2,1,1),
xlim=c(0,10), break.time.by = 1, censor=FALSE)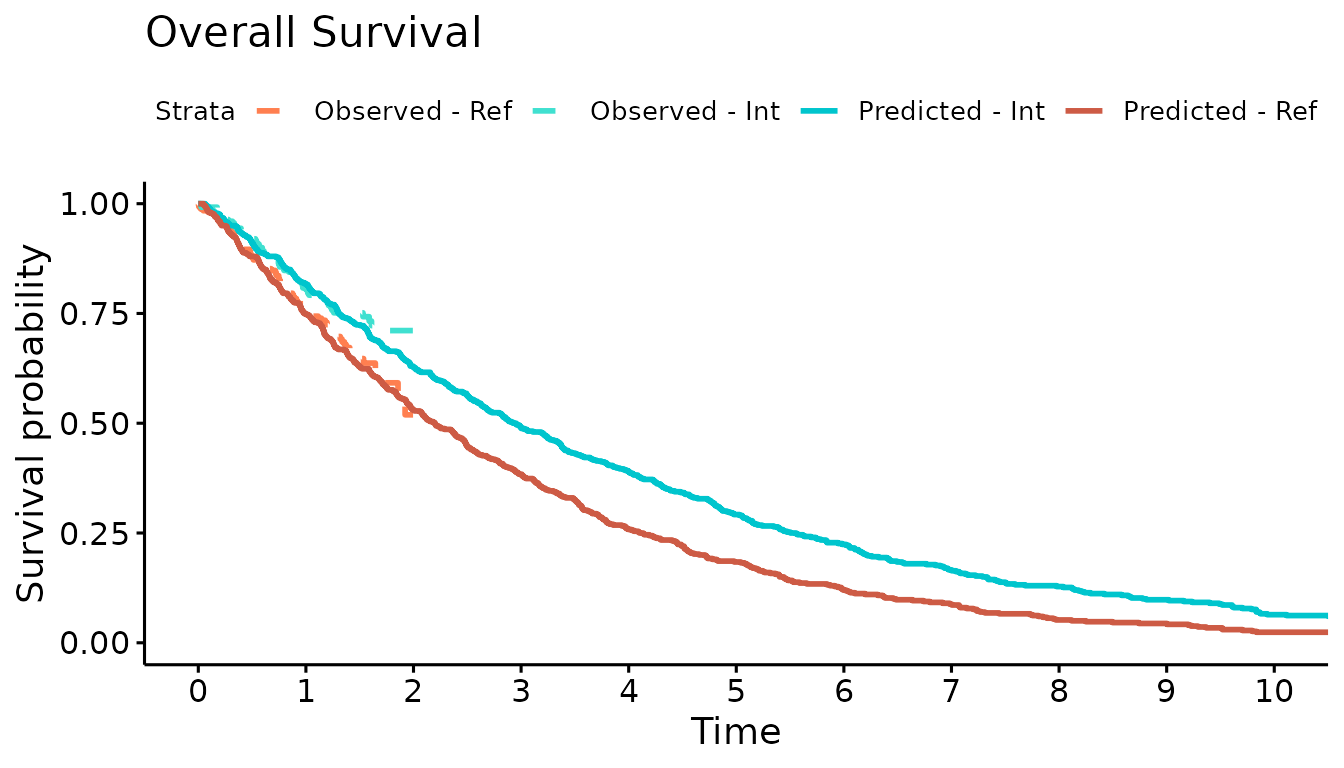
#Progression-free survival
km.est.PFS <- survfit(Surv(AVAL_PFS/365.25, Event_PFS) ~ ARMCD, data = IPD)
KM.PFS.DES <- psa_ipd %>% filter(evtname=="Death") %>% mutate(evttime = ifelse(is.na(pfs.time),evttime,pfs.time),
Event = 1)
sim.km.PFS <- survfit(Surv(evttime, Event) ~ arm, data = KM.PFS.DES)
km.comb <- list(Observed = km.est.PFS,
Predicted = sim.km.PFS)
ggsurvplot(km.comb,combine = TRUE,
title="Progression-free Survival",
palette=c("coral","turquoise","turquoise3","coral3"),
legend.labs=c("Observed - Ref","Observed - Int","Predicted - Int","Predicted - Ref"),
linetype = c(2,2,1,1),
xlim=c(0,5), break.time.by = 1, censor = FALSE)